Visual Abstraction in DNA Nanotechnology
Tutorial on Biological Visualization
Date: Thu, 5 September 2019 at 9.00-10.30
Organizers:
- Haichao Miao (TU Wien, Austria)
- Ivan Barišić (Austrian Institute of Technology, Austria)
Abstract:
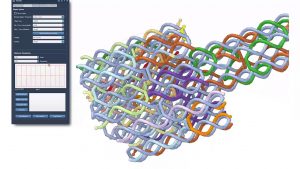
In this tutorial, we describe the challenges, insights, and solutions when developing a visualization system for the in silico design of DNA nanostructures. These nanoscale objects are increasingly abstracted to reduce visual complexity. We describe the abstraction along the aspect of scale and dimensions where a multitude of visual encodings are designed. Seamless transitions depict the perceivable correspondence between these representations. Finally, a provenance-inspired abstraction map guides the scientists to relevant visualizations.
Rapid Prototyping with Inviwo for Medical Applications
Tutorial on Medical Visualization
Date: Thu, 5 September 2019 at 16.00-17.30
Organizers:
- Martin Falk (Linköping University, Sweden)
- Daniel Jönsson (Linköping University, Sweden)
Abstract:
Inviwo is a rapid prototyping framework for visualizing spatial and abstract data. In this tutorial, we show how Inviwo can be utilized for easily creating visualizations in the medical domain. We provide an overview of the concepts used in Inviwo like its visual network editor and the associated data flow paradigm. Several hands-on examples illustrate how to import data and build your own visualization networks. In the second part, we cover how to extend the existing functionality using python scripting and C++. By the end of the tutorial, you will be familiar with Inviwo’s capabilities and use it effectively.
Inviwo is available at https://inviwo.org and Github (https://github.com/inviwo/inviwo).
Machine Learning (ML) Tutorial

Date: Thu, 5 September 2019 at 17.30-19.00
Organizers:
- Timo Ropinski (Ulm University, Germany)
- Pedro Hermosilla Casajus (Ulm University, Germany)
Abstract:
Within this tutorial a basic introduction will be given into machine learning as it is used within visual computing in biology and medicine. The focus will be on convolutional neural networks (CNNs), and their application to image classification and segmentation. For these tasks, current network architectures and training data sets are discussed. After completing the tutorial, attends should be able to use TensorFlow to train their own networks with their own data.
